


QUANTITATIVE FLUORESCENT POLYMERASE CHAIN
REACTION (QFPCR) IN THE PRENATAL AND POSTNATAL
DIAGNOSIS OF THE MOST FREQUENT ANEUPLOIDIES
Macek M Sr1,*, Krebsova A1,2, Brou_ková M1, Matj_ková M1,
Machatková M1, Diblík J1, Sperling K2, Vorsanova S3, Kutsev S4,
Zerova S5, Arbuzova S6, Chudoba D1, Novotná D1
*Corresponding Author: Associate Professor Milan Macek, Sr., MD, PhD, Centre of Reproductive Genetics, Institute of Biology and Medical Genetics, University Hospital Motol, Charles University, 2nd Medical School, V uvalu 84, Prague, CZ 150 06, Czech Republic; Tel.: +4202-2443-3534; Fax: +4202-2443-3525; E-mail: pavel.roubic@lfmotol.cuni.cz
page: 87
|
|
INTRODUCTION
Quantitative fluorescent polymerase chain reaction (QFPCR) represents a rapid and reliable molecular genetic method for the detection of the most frequent aneuploidies of the chromosomes 13, 18, 21, X and Y in prenatal diagnosis that does not require long-term cultivation of fetal cells necessary for classical cytogenetic analysis. Its feasibility was confirmed on amniocytes from genetic amniocentesis [1-8], on chorionic villi samples (CVS) [9], on coelomic cells [10], on fetal cells from cervical/uterine lavages [11-13], and on fetal cells or fetal DNA in maternal blood [14]. This method is also feasible on single cell level in pre-implantation genetic diagnosis (PGD) [3,15,16].
Quantitative fluorescent PCR uses multiplex PC reactions of specific STRs from different regions of the studied chromosomes [17] and fluorochrome labeling of specific STR primers [18,19]. It is based on the pioneering works of von Eggeling et al. [20], Mansfield [21] and Mutter and Pomponio [22]. New, highly polymorphic microsatellite markers provide the highest genetic informativity for the detection of heterochromosomal and autosomal aneuploidies [23-25]. Rahil et al.[26] recently introduced non polymorphic target genes on chromosomes 13, 18 and 21 for QFPCR aneuploidy detection, and Pont-Kingdon and Lyon (27) for allele quantification combined with melting curves analysis of single-nucleotide polymorphism loci. Quantitative fluorescent PCR also assures exact diagnosis of partial chromosome aneuploidies [28,29] in prenatal diagnosis, as well as in solid tumors in children with the detection of loss of heterozygosity.
The amnio PCR minimizes parental anxiety by aneuploidy detection in genetic risk pregnancies with abnormal results of ultrasound and biochemical screening in the first and second trimesters [30-33] with a 99% reliability. Moreover, QFPCR provides easy and reliable detection of parental and meiotic origin of aneuploidies for genetic counseling, as well as for the study of the impact of genetic and exogenous factors on the pathogenesis of non-disjunction [1,34,35].The aim of our report is to summarize the implementation of QFPCR in prenatal and postnatal diagnosis of aneuploidies.
Table 1(a). Applied STR markers. (bp denotes the length in base pairs; repeat describes the number of bases in a tandem repeat of a particular STR; AMX/Y: the repeat field is empty because this marker is not an STR; markers in italics represent new STR markers for chromosomes 13, 18 and 21, introduced and verified in our study.)
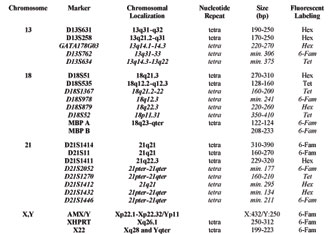
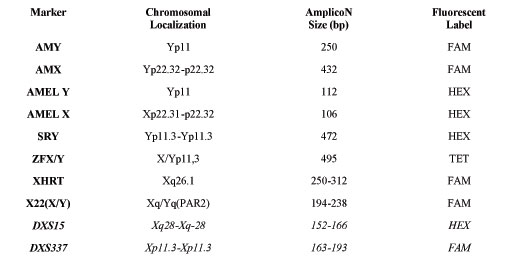
Table 1(b). STR markers used for heterochromosome evaluation. (Markers in italics represent new STR markers, introduced and verified in our study, that allow quantification of chromosome X aneuploidies and increase the proportion of genetic informative families.)
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

