


DETOXIFICATION GENE POLYMORPHISMS AND SUSCEPTIBILITY TO
SPORADIC MOTOR NEURON DISEASE IN THE RUSSIAN POPULATION
Shadrina MI1,*, Slominsky PA1, Zherebtsova AL1, Levitsky GN2, Levitskaya NI2,
Alekhin AV2, Semenova EV1, Serdyuk AV2, Skvortsova VL2, Limborska SA1
*Corresponding Author: Dr. Maria I. Shadrina - Institute of Molecular Genetics, Russian Academy of Scences, Kurchatov sq.2, Moscow 123 182, Russia; Tel.: +7-095-196-0210; Fax: +7-095-196-0221; E-mail: shadrina@ img.ras.ru
page: 31
|
|
RESULTS
CYP2E1 Gene Polymorphism. The results of CYP2E1 genotyping are presented in Table 2. Note that we identified only the common allele, CYP2E1*1C, carrying six repeats in the promoter region, and the insertion allele CYP2E1*1D carrying eight repeats, and did not identify the wild type allele, CYP2E1*1A, which contains only five repeats. The CYP2E1*1D allele was rare, especially in the control group, and there was no difference in the frequency of this allele between our controls (2.5%) and a Caucasian population (2%) [12]. However, a considerably increased frequency of the CYP2E1*1D was observed in MND patients (14% versus 2.5% in controls; p <0.0001). Analysis of genotype distributions did not identify the CYP2E1*1D/CYP2E1*1D genotype in the control group. These results are in accordance with those for a Caucasian population [12]. Genotype distributions showed statistically significant differences between our patients and controls (p = 0.0018). There was observed a considerably increased frequency of CYP2E1*1D homozygotes among our MND patients (see Table 2). The genotype distribution was consistent with Hardy-Weinberg equilibrium. Because genotypes that contain the insertion are associated with higher levels of enzyme activity, we considered CYP2E1*1D heterozygotes (a) and CYP2E1*1D homozygotes (b) as one genotype. The combined CYP2E1 genotype (a + b) was also essentially predominant in our patients when compared with the control group [RR 2,500, 95% confidence interval (CI) 1,159-5,392].
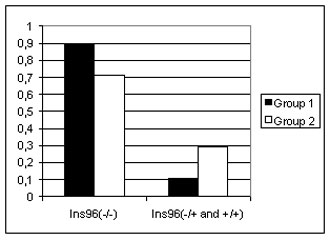
We estimated the possible influence of the CYP2E1*1D allele, with increased enzyme activity, on the clinical expression of MND. To this end, we studied the correlation between the combined CYP2E1 genotype and patients with different clinical features, as listed in Table 1. We found only one cor- relation between clinical diagnosis and the combined geno- type (gamma correlation: Z = 2.838, p = 0.005; Spearmen correlation: R = 0.245, p = 0.033). Further analysis showed that the common allele genotype (CYP2E1*1C/ CYP2E1*1C) is associated with cervical onset ALS, whereas the CYP2E1*1D allele is significantly correlated with other and more malignant forms of MND, such as truncal onset and diffuse onset ALS and progressive bulbar palsy [χ 2 = 3.85, p = 0.0492; see Fig. 1(a)].
CYP2D6 Gene Polymorphism. The CYP2D6 allele and genotype frequencies for our patients and controls are given in Table 3. No significant difference in genotype frequency was observed between our controls and British Caucasian controls [33], the frequency of CYP2D6*4 homozygotes being 4% in both. Neither patient nor control genotype frequencies varied significantly from the Hardy-Weinberg equilibrium.
Although a previous study of ALS reported a considerably increased frequency of the CYP2D6*4 allele [10], a direct χ 2 comparison of the CYP2D6 allele and genotype frequencies in our MND patients and the controls revealed no significant difference (Table 3). The number of CYP2D6*4 homozygotes was elevated in the patient group.
Because only CYP2D6*4 homozygosity results in the PM phenotype [10], we combined the non CYP2D6*4 homozygous (a) and CYP2D6*4 heterozygous (b) genotypes as one genotype. Comparative distribution of the combined CYP2D6 genotype (a+b) and CYP2D6*4 homozygotes revealed no statistically significant difference between our patients and controls (Table 3). CYP2D6*4 homozygotes were more frequent among our patients, but this difference was not statistically significant (p = 0.0754).
We also studied the distribution of the combined CYP2D6 genotype in patients with different clinical features, as CYP2E1 analysis. There was no significant correlation between different clinical features and any genotype of the CYP2D6*4 polymorphism (data not shown).
GSTT1 and GSTM1 Gene Polymorphisms. The genotype frequencies of the GSTT1 and GSTM1 genes observed in our patients and control groups are presented in Table 4. Note that we could conclusively identify only the null genotypes GSTT1(0/0) and GSTM1(0/0). The heterozygotes and homozygotes for the normal allele were combined together and are treated as the GSTT1(+) or GTM1(+) genotypes.
The frequencies of GSTT1 and GSTM1 homozygous deletions in our control group were similar to those reported for Caucasians [20,23,36]. The frequency of GSTT1-null subjects was much lower than that of GSTM1(0/0) genotype and was very similar in both groups (see Table 4). Our results for the GSTT1(0/0) genotype differ from those of Stroombergen et al. [23]. A preponderance of GSTT1-null subjects was observed in the patient group, whereas no association between GSTM1 homozygous deletions and MND were found in the British population [23]. In contrast, we found a significant difference in the GSTM1(0/0) genotype distribution between our patients and controls. We also found significantly reduced GSTM1(0/0) frequency and elevated GSTM1(+) frequency in our patients group (Table 4). We found no significant correlation between the GSTT1-null and GSTM1-null genotypes and different clinical features.
GSTP1 Gene Polymorphism. The results of GSTP1 genotyping are presented in Table 5. The analysis of 13 additional patients did not affect our previously reported results [38]. The wild type allele (GSTP1*A) was the most common allele in the control and patient groups. The frequency of GSTP1*B homozygotes in the control population was similar to that reported for Caucasians [39,40]. The distributions of the GSTP1 genotypes and those predicted by allele frequencies showed that the control and patient groups were in Hardy-Weinberg equilibrium. A slight preponderance of GSTP*B homozygotes was observed among our patients but did not appear to be significant (see Table 5). Because previous studies have demonstrated that the 105Val variant has altered heat stability and catalytic efficiency for various xenobiotics [21], we combined heterozygotes (a) and homozygotes (b) for the GSTP1*B allele into one category. As shown in Table 5, the distributions of the GSTP1 genotype (a+b) and GSTP1*A homozygotes were very similar. We found no significant association between the combined GSTP1 genotypes and the different clinical features listed in Table 1 (data not shown).
Only one correlation between variations in motor neuron involvement and the combined GSTP1 genotypes was found (gamma correlation: Z = 2.909, p = 0.003; Spearmen correlation: R = 0.242, p = 0.036). Further analysis showed that the wild type allele genotype (GSTP1*A/GSTP1*A) was associated with classical upper and lower MN involvement, whereas the presence of the GSTP1*B allele was significantly correlated with predominant lower MN involvement and predominant upper MN involvement [χ 2 = 6.60; p = 0.0102, see Fig. 1(b)].
NAT2 Gene Polymorphism. The three most common slow alleles, S1, S2, and S3 (NAT2*5, NAT2*6, and NAT2*7, respectively) and one wild type fast allele, F1 (NAT2*4), were identified. The number of S1, S2, and S3 alleles and the frequencies of various genotypes assessed separately were rather low (data not shown), so we combined all the alleles into slow (S) and fast (F1), and all the genotypes into rapid acetylators and slow and intermediate acetylators. The frequency of slow and intermediate acetylators was slightly elevated in the MND patient group (57.3%) versus control group (50.5%), were not significant (χ 2 = 0.83, p = 0.36). No significant correlation was observed between acetylator types and clinical features.
Table 2. The allele and genotype distribution for the insertion polymorphism of CYP2E1 gene in MND patients and controls.
|
Allele frequency |
MND patients
N (%) |
Control
N (%) |
2 |
P |
RR (0,95 %CI) |
|
CYP2E1*1C (Ins96(-))
CYP2E1*1D (Ins96(+)) |
129 (86)
21 (14) |
205 (97,5)
5 (2,5) |
15,938a |
<0,0001a |
3,192 (1,445-7,050) |
|
Genotype frequency |
|
|
|
|
|
|
CYP2E1*1C/ CYP2E1*1C
CYP2E1*1C/ CYP2E1*1D (a)
CYP2E1*1D/ CYP2E1*1D (b) |
60 (80,0)
11 (14,7)
4 (5,3) |
100 (95)
5 (5)
0 |
11,571b |
0,0031b |
|
|
CYP2E1*1C/ CYP2E1*1C
a+b |
60 (80,0)
15 (20,0) |
100 (95)
5 (5) |
8,801a |
0,003a |
2,500 (1,159-5,392) |
|
Total number |
75 |
105 |
|
|
|
|
MND - M???r neuron disease,
a - 2 is has been calculated with use 2x2 contingency table, GraphPadInStat program, marked correlation are significant at p<0,05
b - 2 is has been calculated with use of 2x3 contingency table, GraphPadInStat program, marked correlation are significant at p<0,025 according Bonferroni’s correction
|
a)
b)
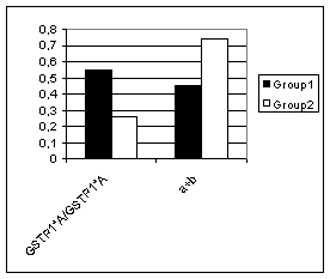
Figure 1. The correlation between clinical features and some gene polymorphisms.
a) The genotype distribution for the insertion polymorphisms of CYP2E1 gene in MND patients with different primary lesion. Group 1 - Cervical onset amyotrophic lateral sclerosis patients; Group 2 – Other motor neuron disease patients.
b) The genotype distribution for Ile105Val polymorphism of GSTP1 gene in MND patients with different disease variants. Group 1 – Motor neuron patients with classical upper and lower MN involvement; Group 2 – Motor neuron patients with predominant lower MN involvement and predominant upper MN involvement. A detail description of GSTP1 Ile105Val polymorphism genotypes see in text.
Table 3. The allele and genotype distribution for the CYP2D6*4 polymorphism in MND patients and controls.
|
Allele frequency |
MND patients
N (%) |
Control
N (%) |
2 |
p |
RR (0,95 %CI) |
|
Non-CYP2D6*4
CYP2D6*4 |
114 (76,0)
36 (24,0) |
174 (82,8)
36 (17,2) |
2,161a |
0,1416a |
1,208 (0,941-1,550) |
|
Genotype frequency |
|
|
|
|
|
|
Non-CYP2D6*4/ Non-CYP2D6*4 (a)
Non-CYP2D6*4/ CYP2D6*4 (b)
CYP2D6*4/CYP2D6*4 |
49 (65,3)
16 (21,3)
10 (13,3) |
74 (70,5)
26 (24,8)
5 (4,7) |
4,247b |
0,1196b |
|
|
a+b
CYP2D6*4/CYP2D6*4 |
65 (86,7)
10 (13,3) |
100 (95,3)
5 (4,7) |
3,161a |
0,0754a |
1,818 (0,879-3,759) |
|
Total number |
75 |
105 |
|
|
|
|
MND - M???r neuron disease,
a - 2 is has been calculated with use 2x2 contingency table, GraphPadInStat program, marked correlation are significant at p<0,05
b - 2 is has been calculated with use of 2x3 contingency table, GraphPadInStat program, marked correlation are significant at p<0,025 according Bonferroni’s correction |
Table 4. The genotype distribution for deletion polymorphism of GSTT1 and GSTM1 genes in MND patients and controls.
|
Genotype frequency |
MND patients
N (%) |
Control
N (%) |
2 |
p |
RR (0,95 %CI) |
|
GSTT1(+)
GSTT1(0/0) |
56 (74,7)
19 (25,3) |
85 (81,0)
20 (19,0) |
0,682a |
0,4090a |
1,208 (0,941-1,550) |
|
GSTM1(+)
GSTM1(0/0) |
45 (60,0)
30 (40,0) |
46 (43,8)
59 (56,2) |
3,963a |
0,0465a |
0,762 (0,593-0,981) |
|
Frequency of genotype combinations |
|
|
|
|
|
|
GSTT1(0/0)/GSTM1(0/0) GSTT1(+)/GSTM1(0/0) GSTT1(0/0)/GSTM1(+)
GSTT1(+)/GSTM1(+) |
8 (10,7)
22 (29,3)
11 (14,7)
34 (45,3) |
16 (15,2)
43 (41)
4 (3,8)
42 (40) |
8,805b |
0,0320b |
|
|
Total number |
75 |
105 |
|
|
|
|
MND - M???r neuron disease,
a - 2 is has been calculated with use 2x2 contingency table, GraphPadInStat program, marked correlation are significant at p<0,05
b - 2 is has been calculated with use of 2x4 contingency table, GraphPadInStat program, marked correlation are significant at p<0,01667 according Bonferroni’s correction |
|
Allele frequency |
MND patients
N (%) |
Control
N (%) |
2 |
p |
RR (0,95 %CI) |
|
GSTP1*A
GSTP1*B |
99 (66)
51 (34) |
146 (69,5)
64 (30,5) |
0,351a |
0,5536a |
1,071 (0,883-1,299) |
|
Genotype frequency |
|
|
|
|
|
|
GSTP1*A/GSTP1*A
GSTP1*A/GSTP1*B (a)
GSTP1*B/GSTP1*B (b) |
31 (41,3)
37 (49,3)
7 (9,3) |
48 (45,7)
50 (47,6)
7 (6,7) |
0,618b |
0,7342b |
|
|
GSTP1*A/GSTP1*A
a+b |
31 (41,3)
44 (58,7) |
48 (45,7)
57 (54,3) |
0,186a |
0,6660a |
1,077 (0,841-1,378) |
|
Total number |
75 |
105 |
Table 5. The allele and genotype distribution for Ile105Val polymorphism of GSTP1 gene in MND patients and controls.
|
|
|
|
MND - M???r neuron disease,
a - 2 is has been calculated with use 2x2 contingency table, GraphPadInStat program, marked correlation are significant at p<0,05
b - 2 is has been calculated with use of 2x3 contingency table, GraphPadInStat program, marked correlation are significant at p<0,025 according Bonferroni’s correction |
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

