


PRIMED IN SITU AND PEPTIDE NUCLEIC ACID:
TWO FAST AND EFFICIENT TECHNIQUES FOR
IN SITU CHROMOSOMAL DETECTION
Pellestor F*
*Corresponding Author: : Dr. Franck Pellestor, Institut de Genétique Humaine, CNRS UPR 1142, 141 rue de la Cardonille, F-34396 Montpellier Cedex 5, France; Tel.: +33-(0)4-99-61-99-12; Fax: +33-(0)4-99-61-99-01; E-mail: Franck.Pellestor@igh.cnrs.fr
page: 61
|
|
THE PNA TECHNIQUE
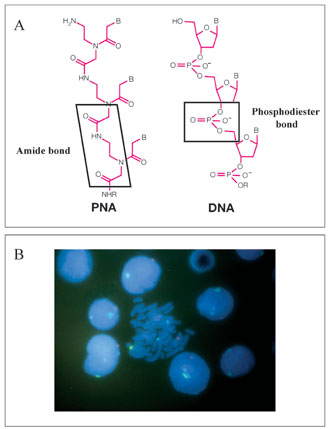
Definition and Properties. Peptide nucleic acids (PNAs) constitute a new class of DNA probes, that provide an interesting alternative to FISH and PRINS. The PNAs are synthetic mimics of DNA in which the deoxyribose phosphate backbone supporting the nucleic acid bases, is replaced by a non-charged peptide backbone [Fig. 2(A)]. The unique chemical makeup of these probes confer a number of beneficial properties, including enhanced hybridization rates, resistance to nucleases and proteases, and the ability to penetrate condensed biological structures. The neutral backbone of PNA provides strong binding between PNA/DNA or PNA/RNA strands and greater specificity of interaction than their DNA counterparts. While they hybridize according to normal Watson-Crick base pairing rules, PNAs have been shown to bind to DNA or RNA targets with higher affinity than the corresponding oligonucleotides. Unlike DNA probes, that require high salt concentration to bind, PNA probes can bind to DNA or RNA under low ionic strength conditions that disfavor re-annealing of complementary strands. Experiments with homopyrimidine strands have shown that the Tm of a 6 mer PNAT/DNAdA was 31°C in comparison to a DNAdt/DNAdA 6-mer duplex that denatures at a temperature of less than 10°C. Experiments done with PNA probes containing all four bases have demonstrated that there is an increase of the Tm of about 1°C per base pair in PNA hybrids compared to DNA/DNA or RNA/ RNA duplexes. In addition, a PNA/DNA mismatch is more destabilizing than a mismatch in a DNA/DNA duplex. A single mismatch in mixed PNA/DNA 15-mer decreases the Tm by 15°C. In the corresponding DNA/DNA duplex, a single mismatch decreases the Tm by only 11°C. Moreover, the use of even shorter PNA probes can further increase PNA specificity. This advantage is particularly important for hybridization with short probes targeting repetitive sequences, because both the length and the repetitive nature of these genomic targets will favor re-naturation over hybridization with probes.
These outlined properties mean that it is not necessary to design long PNA probes. Short PNA oligomers, from17 to 22 bases units, constitute efficient tools for detecting specific DNA sequences with fast hybridization kinetics (20 to 60 min.) over a wide pH range. In addition, PNAs are not limited to any detection procedure. The PNAs can be labeled with a large variety of reporter molecules (enzymes, haptens, fluorophores) [Fig. 2(B)].
Applications and Perspectives. The PNA was originally conceived as reagents for sequence-specific recognition of double-stranded DNA via triple helix formation [26]. However, the unique properties of PNA as a DNA mimic have quickly led to the development of various applications. Most notably, PNAs find current use in molecular biological techniques as specific and sensitive probes for complementary nucleic acids. The PNA oligomers are powerful probes in Southern and Northern blotting but also for mutation research. Polymerization of PNA oligomers into polyacrylamide gels creates a medium in which the unique properties of PNA/DNA interactions are utilized to achieve hybridization with single-stranded target DNA during affinity electrophoresis. Such a procedure has been used to identify single point mutations [27]. However, the greatest benefit of PNA comes from the development of new diagnostic assays. For example, PNA technology drastically reduces assay times compared to standard methods for bacterial analysis, by using PNA probes that target specific ribosomial RNA sequences [28]. Because of their high affinity and stability, PNA oligomers are also employed in transcription inhibition (antigene strategy) and translation inhibition (antisens strategy) [29]. The use of PNA probes in cytogenetics, as an efficient variation to FISH for chromosomal identification, is recent. Firstly, it has been demonstrated that PNA probes are useful for detecting telomere repeat sequences [30] and triplet repeat sequences in myotonic dystrophy cells [31]. The high discriminating ability of PNA probes was evidenced by the detection of inter-spaced centromeric DNA repeats that differed by a single base pair [32], suggesting the further use of PNAs for differentiating chromosomal polymorphisms. The availability of chromosome-specific centomeric PNA probes, directly fluorochrome-labeled, has led to the development of rapid PNA protocols for the in situ detection and enumeration of human chromosomes in metaphases and interphase nuclei. Multicolor PNA experiments were thus reported on cells from normal subjects and patients with numerical abnormalities [33,34]. The procedure was also adapted on human sperm [35]. On this material, PNA gave a similar performance to PRINS, pointing out the advantages of both these techniques over conventional FISH in terms of hybridization kinetics, specificity and accessibility to the targeted sequences. The successful adaptation of the PNA technique on human sperm has proven that these probes could be used on difficult and compact biological material.
The fast hybridization, penetration and discrimination of PNA probes make them valuable tools for in situ chromosomal screening. They have great potential for clinical applications, in particular when chromosomal identification must be performed on limited amounts of material, and in a limited time period such as in pre-implantation chromosomal diagnosis.
Figure 2. Peptide nucleic acid labeling. (A) Comparison of PNA and DNA structures. Whereas the DNA involves a phosphate backbone supporting the nucleic acid bases, the PNA backbone is made from repeating N-(2-aminoethyl)-glycine units linked by peptide bonds. The acid nucleic bases are linked to the backbone by methylene carbonyl linkages. (B) Example of PNA labeling on human lymphocytes, using specific-centromeric PNA probes for chromosome 1 (blue signal), chromosome X (red signal) and chromosome Y (green signal).
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

