


PRIMED IN SITU AND PEPTIDE NUCLEIC ACID:
TWO FAST AND EFFICIENT TECHNIQUES FOR
IN SITU CHROMOSOMAL DETECTION
Pellestor F*
*Corresponding Author: : Dr. Franck Pellestor, Institut de Genétique Humaine, CNRS UPR 1142, 141 rue de la Cardonille, F-34396 Montpellier Cedex 5, France; Tel.: +33-(0)4-99-61-99-12; Fax: +33-(0)4-99-61-99-01; E-mail: Franck.Pellestor@igh.cnrs.fr
page: 61
|
|
THE PRINS TECHNIQUE
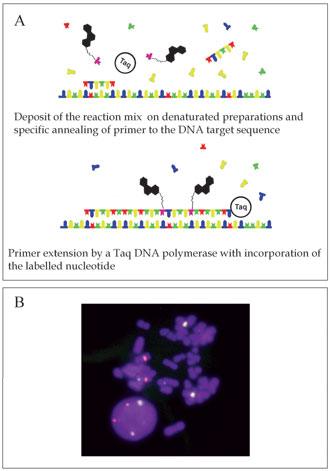
Principles and Methodology. Based on the use of chromosome-specific primers, the PRINS reaction combines the high sensitivity of the polymerase chain reaction (PCR) with the cytological localization of DNA sequences [3]. The chromosomal identification is performed by in situ annealing of specific and unlabeled oligonucleotide primers to complementary sites on denatured chromosome spreads, nuclei or tissue sections. Cells or tissues samples are fixed and denatured before PRINS reaction, both to preserve morphology and to permit access of the reagents to the sequence target. The annealed primers provide initiation sites for chain elongation catalyzd by a Taq DNA polymerase in the presence of free nucleotides (nts), of which at least one is labeled. The in situ visualization of generated fragments results from the incorporation of the labeled nt [Fig. 1(A)].
Primed in situ labeling of human chromosomes is obtained using primers for repeated DNA sequences. An advantage of primers is their ability to differentiate between closely related sequences. This feature has been utilized for generating chromosome-specific primers from the alpha-satellite DNA motif. These centromeric repeats are made up of a variable number of monomeres of 171 bp in length and are organized as alpha-satellite subfamilies. The DNA sequences of momomeres slightly deviate among subfamilies and individual chromosomes. The chromosome specificity of PRINS labeling is based on the use of primers generated from these chromosome-specific alpha-satellite DNA sequences. The lengths of the PRINS primers range from 18 to 35 nts. Compared to the size of DNA repetitive probes (250 to 600 bp), this small size greatly facilitates their in situ accessibility to their genomic target sequences. This is particularly significant in cells with highly condensed nuclei, such as spermatozoa. Because they are unlabeled, high amounts of primers can also be used in PRINS reactions without inducing background signals. The complementation process between the primer and its centromeric target will be so specific that a simple mismatch between the 3’-end of the primer and the genomic sequence will prevent initiation of the in situ elongation by the Taq DNA polymerase [4]. Thus, it has been possible to define specific alpha-satellite primers for some chromosomes undistinguishable by FISH with centromeric probes, such as chromosomes 13 and 21, which share 99.7% homology in their alpha-satellite DNA sequences [5].
Initially, PRINS reactions were performed either on a hotplate or in a water bath, but these procedures did not allow precise and durable temperature control. The PRINS protocol has been considerably improved and simplified by the introduction of programmable temperature cyclers equipped with a flat plate block. The use of automatic thermocyclers allows an optimization of both annealing and extension conditions. Thus, semi-automatic PRINS protocols have been developed offering a high reproducibility in labeling reaction.
An additional improvement was the direct use of fluorochromes in sequential PRINS reactions [6,7]. Recently, a new multicolor PRINS protocol has been reported, allowing the performance of ultra-rapid detection on several chromosomes, only by mixing the different fluorochromes during the chain elongation reaction [8]. Each PRINS reaction consists of a unique 4 min. step for annealing and elongation of each chromosome-specific primer. This new sequential procedure simplifies the PRINS technique and provides an easy way to carry out multicolor labeling [Fig. 1(B)].
Applications and Perspectives. The PRINS procedure combines several features that make it very attractive for a number of cytogenetic purposes. Various applications of PRINS have already been developed in humans, mammalians [9,10], fishes [11], insects [12] and plants [13,14], demonstrating that PRINS could easily be adapted to various types of cells.
In humans, the PRINS method has successfully been tested for the assessment of aneuploidy in lymphocytes, amniocytes [15,16] and pre-implantation embryos [17,18]. The use of PRINS has also been reported for analysis of structural aberrations such as translocations, maker chromosomes and ring chromosomes [19]. Since the PRINS reaction with Alu primers give high quality R-like banding on human chromosomes, the procedure has been adapted for the cytogenetic screening of somatic hybrid cell lines and identification of euchromatin in aberrant short arms of acrocentric chromosomes and small ring chromosomes [20].
More recently, the PRINS protocol has been tested to detect fetal cells among separated maternal nucleated cells from peripheral venous blood of 15 pregnant women with singleton pregnancies ranging from 12 to 35 weeks’ gestation [21]. Further applications of PRINS have also been found in tumoral cytogenetics, opening new possibilities for cytogeneticits and pathologists. All these applications point out the potential efficiency of the PRINS method for diagnostic use.
Some research studies have reported the use of PRINS for the direct estimation of disomy and diploidy rates in human sperm [22]. The assessment of aneuploidy rate in human gametes is of critical importance because non disjunctions make a major contribution to the chromosomal abnormalities found in humans. The FISH strategy using centromeric probes was first adapted to ejaculated human spermatozoa. However, FISH analysis of male gametes is hampered by the strong condensation of sperm nuclei. To date, FISH studies of human sperm have reported considerable variations in the frequency of chromosome disomy. In the PRINS reaction, the decondensation of the sperm head is a less limiting factor than in FISH because of the small size of the oligonucleotide primers. This greatly facilitates their penetration into sperm nuclei resulting in a more homogeneous and more rapid labeling of sperm nuclei. Using the PRINS method, diploidy and disomy frequencies have been estimated for 15 autosomes and sex chromosomes in sperm samples from several normal fertile donors. The PRINS procedure has also been used to directly investigate in sperm the meiotic segregation patterns of reciprocal translocations [23].
New improvements of the PRINS are presently ongoing. Thus, the adaptation of the PRINS technique to the in situ detection of unique sequences remains an important challenge. This approach would allow us to do away with tedious protocols for preparing probes, and also to rapidly map newly discovered genes on chromosomes by using synthetic oligonucleotides derived from sequenced DNA. The advantage of the PRINS approach is that it does not rely on the possession of a cloned probe for the target. As long as the sequence of the gene is known, oligonucleotides can be synthesized for use as primers. Several teams have been working on this project. Preliminary results have been obtained on porcine chromosomes [24], and recently efficient PRINS localizations of several genes have been reported on human metaphases [25]. This opens new and promising perspectives for PRINS in the area of physical mapping.
Figure 1. Primer in situ labeling. (A) Principles of the PRINS reaction. (B) Example of three-color PRINS labeling on human lymphocytes. Chromosomes 1, 7 and 9 are labeled in yellow, green and red, respectively.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

