


PRE-IMPLANTATION GENETIC DIAGNOSIS FOR
β-THALASSEMIA, SICKLE CELL SYNDROMES
AND CYSTIC FIBROSIS IN GREECE
Traeger-Synodinos J1,*, Vrettou C1, Tzetis M1, Palmer G2,
Davis S3, Mastrominas M3, Kokali G4, Pandos K4, Kanavakis E1
*Corresponding Author: : Dr. Joanne Traeger-Synodinos, Medical Genetics, Athens University, Choremio Research Laboratory, St. Sophia’s Children’s Hospital, Thivon and Levadias Streets, Athens 11527, Greece; Tel.: +30-210-746-7461; Fax: +30-210-779-5553; E-mail: jtraeger@cc.uoa.gr
page: 25
|
|
PRE-IMPLANTATION GENETIC DIAGNOSIS FOR CYSTIC FIBROSIS
Cystic fibrosis is one of the most common genetic diseases targeted for prevention programs. It is caused by defects (>1,000 reported to date) in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (250 kb), consisting of 27 exons and located on chromosome 7q31-32 [22]. Defects in the CFTR protein cause abnormal chloride concentration along the apical membrane of epithelial cells resulting in progressive lung disease, pancreatic dysfunction and male infertility. Infertile men who are otherwise asymptomatic, have an increased chance of having CFTR mutations [most commonly those with congenital bilateral absence of the vas deferens (CBAVD)] [21,23]. Thus, couples seeking assisted reproduction due to male infertility may have a markedly increased risk of initiating a CF-affected pregnancy; therefore, PGD is also indicated for such couples.
In Southern Europe, including Greece, CF is characterized by great allelic heterogeneity compared to the populations of Northern Europe [7,24-26]. Most of the approaches for PGD of CF that have been described to date either have not addressed the problem of diagnosing a wide spectrum of mutation combinations [27-31), or require the design of case-specific protocols each time [32]. Protocols based on analysis of CFTR gene linked polymorphic markers have been described that are quite widely applicable for some Northern European populations (33,34), although their usefulness has not been assessed elsewhere.
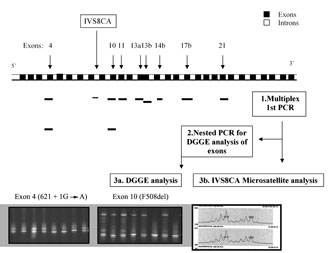
To apply PGD to CF we developed a flexible multiplex PCR protocol, allowing analysis of sequence variations in any combination amongst seven CFTR gene exons which include the majority of Greek CF mutations (exons 4, 10, 11, 13 in two parts, 14b, 17b and 21,) by nested-PCR and DGGE analysis, along with a fluorescently labeled intragenic micro-satellite (IVS8CA) (Fig. 2). Pre-clinical experiments were carried out on 390 single lymphocytes from three CF patients, one heterozygote and one non CF individual, all with known CFTR genotypes. Exons were amplified with a PCR efficiency ranging from 90-100%, with ADO from 0-3.8%. The micro-satellite IVS8CA was co-amplified with an overall PCR efficiency of 92.4 and 10.8% ADO. The method overcomes the need of separate assays for each CFTR gene mutation. Additionally, it facilitates analysis of any informative linked polymorphic sequence variation (within the seven exons), along with micro-satellite useful for minimizing misdiagnosis and for indirect diagnosis. The protocol is robust and flexible for diagnosing diverse CF genotype combinations in single cells, although, to date, we have not completed any clinical PGD cycles for CF [35].
Figure 2. The PGD strategy for analysis of CFTR gene mutations. 1) Multiplex first PCR for appropriate CFTR gene exons (according to parental mutations) and IVS8CA micro-satellite; 2) nested PCR for DGGE analysis of appropriate exons; 3a) DGGE analysis, and 3b) IVS8CA micro-satellite analysis.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

