


WHOLE GENOME ANALYSIS BY ARRAY-BASED
COMPARATIVE GENOMIC HYBRIDIZATION IN
PATIENTS WITH CONGENITAL MALFORMATIONS
Dimova I1, Vazharova R1, Nikolova D1, Tincheva R2,
Nesheva D1, Uzunova Y3, Toncheva D1,*
*Corresponding Author: Professor Draga Toncheva, Department of Medical Genetics, Medical
University Sofia, 2, Zdrave str., 1431 Sofia, Bulgaria; Tel./Fax: +359-2-952-0357; E-mail:dragatoncheva@yahoo.com
page: 33
|
|
RESULTS
We studied blood from our four patients by the method of array-CGH with CytoChip (BlueGnome), covering all autosomes and sex chromosomes at a mean density of 1 BAC clone/0.5 Mb. More than 95% of genomic clones were successfully hybridized in each case. Standard deviation in log2 ratios of Cy3 and Cy5 intensities [test (T) and normal (N) DNA, respectively] ranged between 0.03 and 0,07, depending on the quality of DNA. We used two approaches to identify BACs that showed significant loss or gain in the analyzed samples: a) observation of high-level loss (log2 T:N ratio <–0.5) and of high-level gain (log2 T:N ratio >+0.5) and b) detection of at least one additional adjacent clone with the same aberration in the same probe. The single aberrant clones were excluded from analysis. Using the data base of Blue Fuse (BlueGnome) we determined the copy number poly morphisms in the patients.In patient 1, we found no specific micro-abnormality, but discovered a polymorphism in the following loci: 2p25.3, 4p15.1, 10q11.21, 10q26.3, 11q13.4, 16p12.1, 17q21.31, 19p13.2, 19q13.33 and Yp11.2 (Figure 1). Their population frequency varied between 1-60% according to the BlueGnome data base.In patient 2, no specific abnormalities were found but seven loci showed a variable number of gene copies (polymorphic regions) in the following positions: 1p13.2, 1q21.3, 1q44, 2p16.1, 3q29, 4p16.1 and 5q13.2. In patient 3, array-CGH analysis revealed genomic loss in the long arm of chromosome 18, which, on the basis of the value of log2 ratio, was interpreted as a mosaic form (Figure 2). The deletion comprised a DNA sequence from 18q21.1 to 18q23 (about 28 Mb) (Figure 3). Beside the deletion, we identified gene copy polymorphisms in positions 8p23.1, 10q26.3 and Yp11.2 On the basis of the presence of the large deletion 18q21-23, we repeated cyto genetic analyses in 100 cells and found a low frequency of mosaicism: 46, XY (83%)/46,XY,del (18)(q21-qter) (Figure 4).In patient 4, we established copy number polymor phisms in the loci 1p31.1, 4p16.1 and 8p23.2.
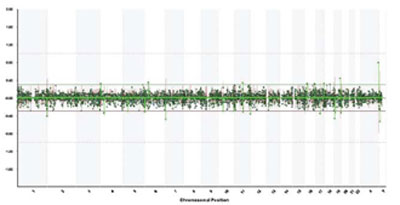
Figure 1. Genomic profile across all chromosomes in patient 1. X-axis: genomic clones on chromosomes 1-22, X and Y; Y-axis: log2 ratio of test to normal DNA for each clone.
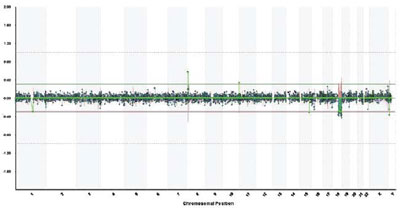
Figure 2. Genomic profile across all chromosomes in patient 3, showing deletion 18q. X-axis: genomic clones on chromosomes 1-22, X and Y; Y-axis: log2 ratio of test to normal DNA for each clone.
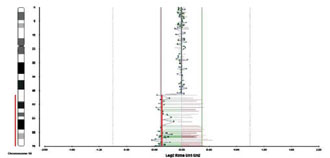
Figure 3. Genomic profile of patient 3 for chromosome 18, revealing deletion of 18q21-23. X- log2 ratio of test to normal DNA for clones on chromosome 18; Y-axis:geno mic clones of chromosome 18.

Figure 4. Karyotype of patient 3. A: normal cell line 46,XY; B: abnormal karyotype with the 18q deletion.
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

