


A STRATEGY FOR THE CHARACTERIZATION OF SMALL
SUPERNUMERARY MARKER CHROMOSOMES (SMC)
Liehr T*, Nietzel A, Weise A, Mrasek K, von Eggeling F, Claussen U, Starke H
*Corresponding Author: Dr. Thomas Liehr, Institut für Humangenetik, Postfach, D-07740 Jena, Germany; Tel: +49-3641-935533; Fax. +49-3641-935502; E-mail: i8lith@mti.uni-jena.de
page: 69
|
|
RESULTS AND DISCUSSION
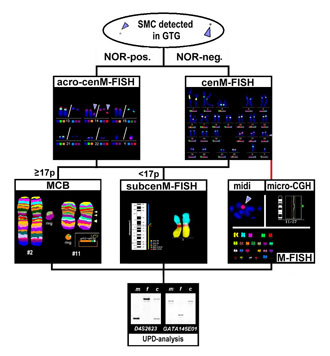
The detection of an SMC is nearly always unexpected by the clinician, and more or less an accidental result in cytogenetics. To facilitate the characterization of SMC according to the novel molecular-cytogenetic possibilities, we here propose a strategy for SMC-characterization (see Fig. 1).
Supernumerary marker chromosomes are initially detected in a GTG-banding analysis and can be characterized for the presence of an acrocentric-chromosome-derived short arm by NOR-staining. According to the NOR-staining result (NOR-negative or NOR-positive) the origin of the SMC can be determined by application of all human centromeres in one experiment (i.e., cenM-FISH [8]) or by the acrocenM-FISH probe set [9], respectively. These approaches have been successfully used in about 100 cases with SMC [8,13-14,18-22; unpublished data].
In general, neocentromeric SMC are not stained by any of the two aforementioned approaches; in this case other approaches for their characterization, such as M-FISH [17], microdissection of the SMC [e.g., 23] or micro-CGH [23], have to be performed. About 60 cases have been reported up to present in the literature with neocentromeres (for review see [24]).
After determination of the origin of a small SMC the most important question to address is, is there euchromatic material of the corresponding chromosome on it, or is there not? This question for partial trisomy can be studied by hybridizing whole chromosome painting (WCP) or chromosome arm specific probes [e.g., 22]. Another possibility is the use of probes that are able to subclassify chromosomal regions [e.g., 23] or MCB [12,14,17]. At present, we are working on a probe set specific for the peri-centric region of all human chromosomes, the subcenM-FISH technique [10]. In the strategy proposed in Fig. 1, MCB is suggested for the characterization of the euchromatic content of SMC larger than 17p and subcenM-FISH for the smaller ones (see Fig. 1).
Various mechanism of formation of small SMC have been proposed: trisomy rescue, monosomy rescue, post fertilization errors and gamete complementation. Recently, another mechanism was proposed, which speculates that SMC are derived from a transfection of the chromosome into the zygote derived from superfluous haploid pronucleus that is usually degraded by deoxyribonucleases or other means [25]. Meta analysis of SMC associated with UPD revealed functional trisomy rescue as the most likely mechanism in marker chromosome formation [5]. The underlying mechanism is always two events: either two meiotic, or one meiotic and one mitotic, which lead to the inheritance of both homologous chromosomes from only one parent. Uniparental disomy has been reported for the majority of human chromosomes [4,26]. Therefore, in the case of identification and characterization of a small SMC, an exclusion of the UPD of the two normal sister chromosomes by molecular genetic methods is reasonable [13,18,22].
In summary, as none of the available approaches can, for technical reasons, ever be fully informative by itself, the cytogenetist always has to combine the available methods, depending on the problem to be addressed. GTG-banding is still the gold standard, which as well the starting point for any molecular cytogenetic for the characterization of small SMC; acrocenM-FISH characterizes its origin, MCB or subcenM-FISH gives information on additional euchromatin and molecular genetics uncovers eventually present UPDs. The strategy proposed here (Fig. 1) might be changed again, depending on further developments of available methods, or an increase in our knowledge of SMC.
Figure 1. We here propose a strategy for SMC-characterization: after initial detection of a SMC in GTG-banding analysis and its characterization for the presence of a NOR-positive region, the origin of the SMC can be clarified by either by application of cenM-FISH [8] (NOR-negative cases) or acrocenM-FISH [9] (NOR-negative cases). To exclude additional euchromatic material of the corresponding chromosome on the SMC, MCB can be performed successfully in the case of an SMC equal or larger in size than 17p [12,14,17]. If smaller, this test should be done by centromere-near locus-specific probes, e.g., in the subcenM-FISH probe-set [10]. If the SMC gave no specific signals in cenM-FISH, a neocentromeric SMC must be suspected; in this case, other approaches for its characterization, such as M-FISH [e.g., 17], micro-dissection of the SMC [e.g., 23] or micro-CGH [e.g., 23] have to be done. Finally, exclusion of UPD in the case of identification and characterization of a small SMC by molecular genetic methods is necessary [13,18,22].
|
|
|
|



 |
Number 27
VOL. 27 (2), 2024 |
Number 27
VOL. 27 (1), 2024 |
Number 26
Number 26 VOL. 26(2), 2023 All in one |
Number 26
VOL. 26(2), 2023 |
Number 26
VOL. 26, 2023 Supplement |
Number 26
VOL. 26(1), 2023 |
Number 25
VOL. 25(2), 2022 |
Number 25
VOL. 25 (1), 2022 |
Number 24
VOL. 24(2), 2021 |
Number 24
VOL. 24(1), 2021 |
Number 23
VOL. 23(2), 2020 |
Number 22
VOL. 22(2), 2019 |
Number 22
VOL. 22(1), 2019 |
Number 22
VOL. 22, 2019 Supplement |
Number 21
VOL. 21(2), 2018 |
Number 21
VOL. 21 (1), 2018 |
Number 21
VOL. 21, 2018 Supplement |
Number 20
VOL. 20 (2), 2017 |
Number 20
VOL. 20 (1), 2017 |
Number 19
VOL. 19 (2), 2016 |
Number 19
VOL. 19 (1), 2016 |
Number 18
VOL. 18 (2), 2015 |
Number 18
VOL. 18 (1), 2015 |
Number 17
VOL. 17 (2), 2014 |
Number 17
VOL. 17 (1), 2014 |
Number 16
VOL. 16 (2), 2013 |
Number 16
VOL. 16 (1), 2013 |
Number 15
VOL. 15 (2), 2012 |
Number 15
VOL. 15, 2012 Supplement |
Number 15
Vol. 15 (1), 2012 |
Number 14
14 - Vol. 14 (2), 2011 |
Number 14
The 9th Balkan Congress of Medical Genetics |
Number 14
14 - Vol. 14 (1), 2011 |
Number 13
Vol. 13 (2), 2010 |
Number 13
Vol.13 (1), 2010 |
Number 12
Vol.12 (2), 2009 |
Number 12
Vol.12 (1), 2009 |
Number 11
Vol.11 (2),2008 |
Number 11
Vol.11 (1),2008 |
Number 10
Vol.10 (2), 2007 |
Number 10
10 (1),2007 |
Number 9
1&2, 2006 |
Number 9
3&4, 2006 |
Number 8
1&2, 2005 |
Number 8
3&4, 2004 |
Number 7
1&2, 2004 |
Number 6
3&4, 2003 |
Number 6
1&2, 2003 |
Number 5
3&4, 2002 |
Number 5
1&2, 2002 |
Number 4
Vol.3 (4), 2000 |
Number 4
Vol.2 (4), 1999 |
Number 4
Vol.1 (4), 1998 |
Number 4
3&4, 2001 |
Number 4
1&2, 2001 |
Number 3
Vol.3 (3), 2000 |
Number 3
Vol.2 (3), 1999 |
Number 3
Vol.1 (3), 1998 |
Number 2
Vol.3(2), 2000 |
Number 2
Vol.1 (2), 1998 |
Number 2
Vol.2 (2), 1999 |
Number 1
Vol.3 (1), 2000 |
Number 1
Vol.2 (1), 1999 |
Number 1
Vol.1 (1), 1998 |
|
|

